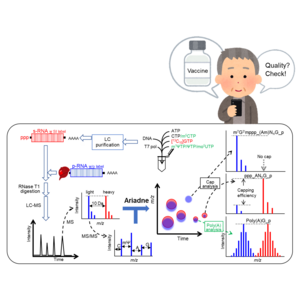
Image: mRNA molecules (p-RNA) were mixed with stable isotope (SI) labeled standards (s-RNA) before digestion and using a combination of liquid chromatography and mass spectrometry (LC-MS). can be characterized as The output is analyzed using Ariadne software to provide information on sequence, ‘cap’ status and tail length. This provides important information about the quality of mRNA pharmaceuticals.
opinion more
Credit: Tokyo Metropolitan University
Tokyo, Japan – Researchers at Tokyo Metropolitan University and RIKEN CSRS have developed a new analytical platform based on liquid chromatography, mass spectrometry, and software analysis to quantify the structure of messenger RNA (mRNA)-based pharmaceuticals . They can elucidate mRNA sequences while quantifying the ‘capping’ of one end of the molecule and the tail integrity of the other end. All of these are essential for mRNA pharmaceuticals. This method is an important innovation for quality control in production lines.
Messenger RNA (mRNA) is the key molecule that transfers the sequence information stored in DNA to the ribosome, the protein-producing machinery of the cell. However, mRNA does not necessarily have to be derived from DNA. By artificially designing mRNA molecules, they can be used as drugs that enable cells to produce specific protein structures. It is like an antigen that triggers an immune response and acts as a vaccine. It has many advantages over competing technologies. It is considered much safer as it does not integrate into the host cell genome. They are relatively easy to manufacture and scale up. The technology is now showing promise in treating a variety of other diseases, including cancer and metabolic disorders.
However, the widespread adoption of this new technology necessitated a way to perform quality control in a more effective and efficient manner. There are three key components to mRNA medicines. A sequence that determines which proteins are synthesized. 5′-capping allows efficient reading of the mRNA during protein translation. A poly(A) tail that weakens the immune response to the foreign mRNA itself. All three must be functioning properly for treatment to be effective. However, there is currently no way to quantify all three states at once.
A team led by Dr. Masato Taoka of Tokyo Metropolitan University and Hiroshi Nakayama of RIKEN CSRS has developed an analytical platform that combines liquid chromatography, mass spectrometry and automated software analysis to quantitatively monitor the properties of mRNA molecules. Did. Teams’ platform combines two key innovations. First, we use liquid chromatography and mass spectrometry to systematically compare various fragments of the tested mRNA molecule to a similarly fragmented reference mRNA labeled with a stable carbon isotope. Second, automated analysis using Ariadne software helps confirm structures with the help of sequence databases. The team found that their analysis platform successfully assigned a reference primary structure and was able to rapidly identify even the slightest changes in the tested mRNA molecules. At the same time, quantitative information on capping and tail groups was also obtained.
The method is applicable to a wide range of mRNA lengths and sequences from disparate sources, allowing all three parts to be analyzed at once. This promises unrivaled efficiency in checking the quality of mRNA pharmaceuticals, both currently used and yet to be developed.
journal
analytical chemistry
article title
Liquid Chromatography-Mass Spectrometry-Based Qualitative Profiling of mRNA Therapeutics Using Stable Isotope Labeled Standards Followed by Automated Quantification Software Ariadne
Article publication date
December 27, 2022
Disclaimer: AAAS and EurekAlert! EurekAlert! is not responsible for the accuracy of news releases posted. Use of information by contributors or via the EurekAlert system.